Sentinel cards found to be effective for indoor SARS-CoV-2 monitoring

In a recent study posted to the bioRxiv* preprint server, a team of researchers used sentinel cards for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) monitoring in schools where individual-level sampling is tedious due to privacy or compliance reasons and higher cost.
Study: Sentinel Cards Provide Practical SARS-CoV-2 Monitoring in School Settings. Image Credit: FOTOGRIN/Shutterstock
There is a higher risk of SARS-CoV-2 transmission in indoor settings, such as school premises. Although convenient for some, not every child has access to a quiet learning environment and stable internet connectivity. Therefore, health authorities recommended a return to in-person learning from remote learning to restore education equality, provided it is safe, for which effective SARS-CoV-2 monitoring is a pre-requisite. However, the labor cost and difficulties in reporting results to health authorities make daily nasal swab testing to monitor the SARS-CoV-2 spread difficult.
The authors of this study have previously implemented environmental monitoring strategies that effectively circumvented clinical swabs to demonstrate that viral signals from COVID-19 patients in indoor environments accumulate on high-touch surfaces and floors. Additionally, the SARS-CoV-2 RNA persists for weeks on indoor surfaces. Thus, there is a need to establish an effective post-sampling cleaning procedure that determines whether consecutive SARS-CoV-2-positives are from new deposition events and when an infected individual contacted a surface.
Thus, there is a need to establish an effective post-sampling cleaning procedure that determines whether consecutive SARS-CoV-2-positives are new.
About the study
In the present study, researchers used 100 cm2 laminated cards to prepare mock sentinel surfaces to help determine whether a SARS-CoV-2-infected individual was recently present in an indoor space. These sentinel surfaces were inoculated with 10 μL of a dilution series of heat-inactivated SARS-CoV-2 WA-1 strain and then wiped with a cleaning solution for five days. In addition, the researchers collected swabs for testing from the sentinel card surfaces at three points, before and after inoculation and after wiping/cleaning.
The researchers used a factorial study design, wherein they used two transport media, three cleaning methods, and three viral load concentrations (high, medium, low) to test the sentinel cards. They performed each condition in triplicate for 54 cards and then a three-step swabbing process on each card over five days.
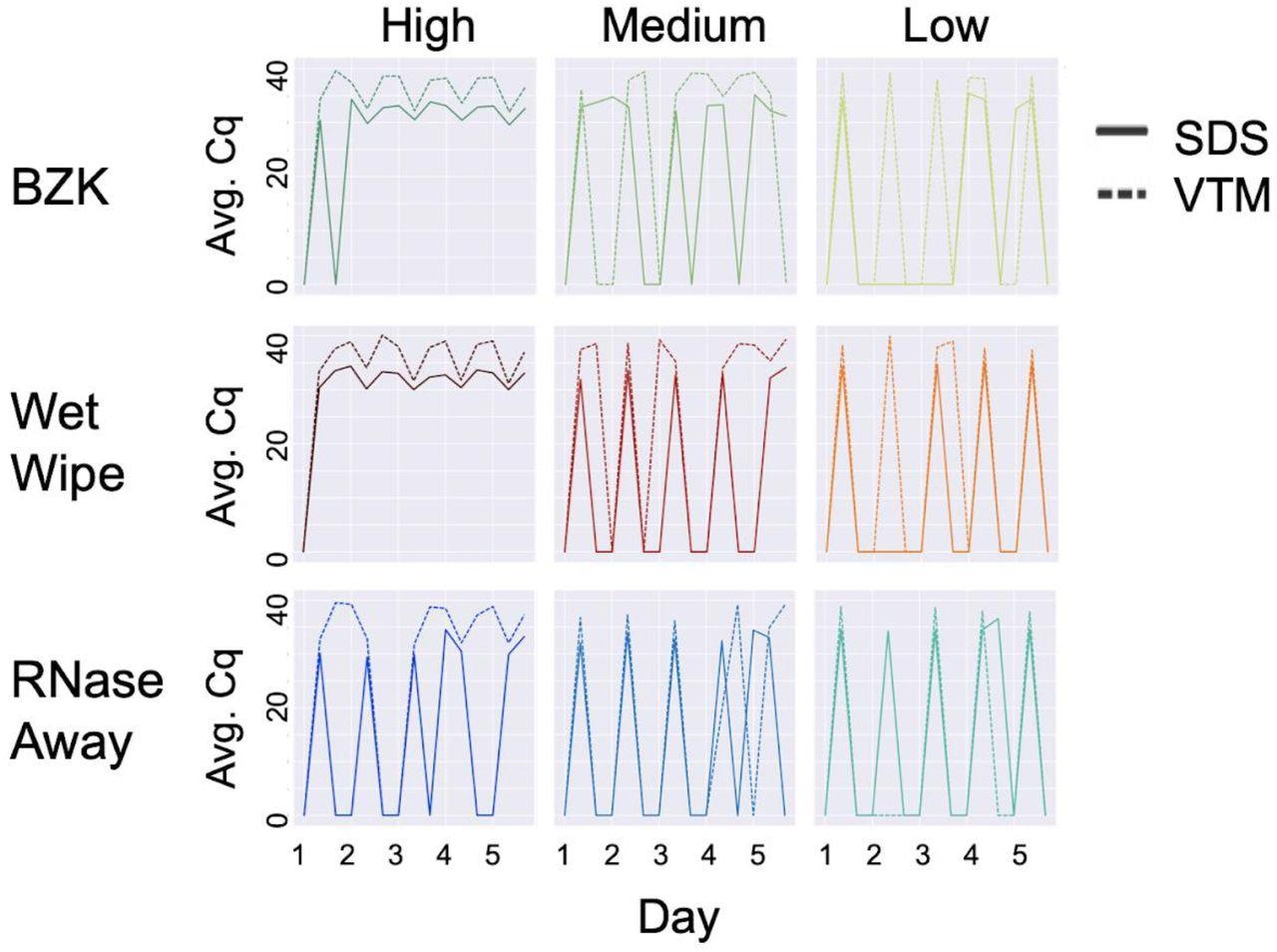
Effect of cleaning solution at high, medium and low viral load with different swabbing media. On each day, three samples were taken: (1) before addition of viral particles, (2) after addition, and (3) after cleaning. Therefore, the expected pattern is a train of 5 spikes, starting at zero, rising to the maximum Cq value, returning to zero the same day, and staying at zero until the next day, as seen for SDS in the low load condition with RNase away (bottom right panel, solid lines). High, medium, and low viral load were defined as (4 x 104), (1 x 104), and (2.5 x 103), respectively. Average Cq (Avg. Cq) was calculated as a mean Cq value from three samples. Two viral transport media were tested: SDS (0.5% w/v sodium dodecyl sulfate (SDS) and VTM (Viral Transport Medium). Effective cleaning reset Cq for each day. RNase away was shown to be effective at each viral load, whereas benzalkonium chloride (BZK) and wet wipes were only effective at medium and low viral load.
The 0.5% w/v sodium dodecyl sulfate (SDS) and Viral Transport Medium (VTM) were used for swabbing, and benzalkonium chloride (BZK) antiseptic towelettes, moist wet wipe (WW) towelettes, and paper towels moistened with RNase AWAY (RA) were used for cleaning.
As measured by droplet digital PCR, high, medium, low viral load dilutions were 4 x 104, 1 x 104, 2.5 x 103, and these three dilutions of SARS-CoV-2 viral genomic equivalents represented typically observed ranges of viral loads in school classrooms.
The first swab was collected from each card at the start of the day, then a second swab was collected post addition of the viral spike-in, and the final swab was collected, post wiping the card with the cleaning solution.
The researchers processed VTM and SDS samples using the Perkin-Elmer and the Thermo pipeline, respectively, and then they conducted extraction and quantitative reverse-transcriptase chain reaction (RT-qPCR).
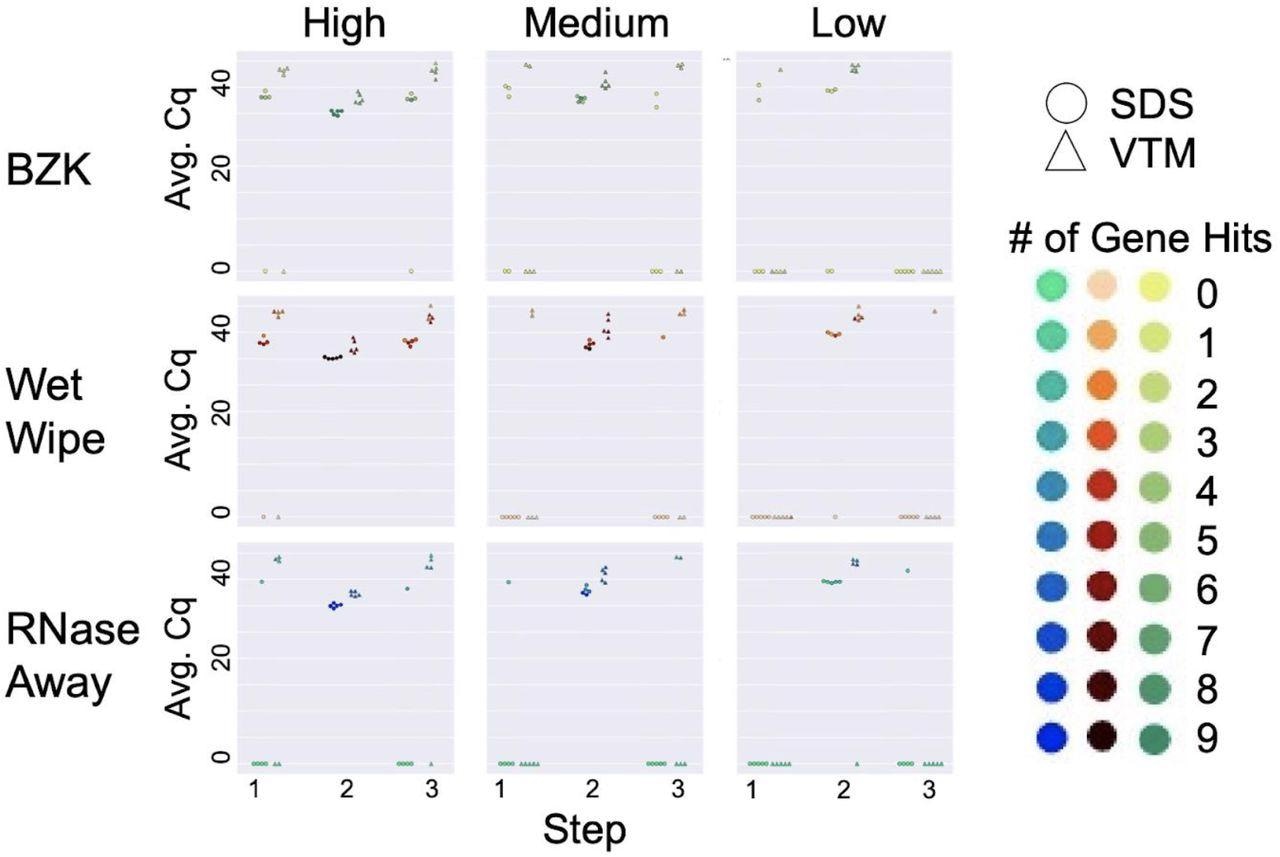
Cleaning solution efficiency after deliberate addition of viral load. Sampling was performed in three steps: initial virus amount (blank) was sampled from the wall for Step 1. Virus was deliberately loaded on the surface and sampled for Step 2. The surface was cleaned with different cleaning methods and sampled for qPCR analysis for Step 3. High, medium, and low viral load were defined as (4 x 104), (1 x 104), and (2.5 x 103), respectively. Average Cq (Avg. Cq) was calculated as a mean Cq value from three samples. Two viral transport media were tested: SDS (0.5% w/v sodium dodecyl sulfate (SDS) and VTM (Viral Transport Medium). Effective cleaning reset Cq for each day (steps 1 and 3), whereas ineffective cleaning retained high viral load (non-zero Cq) at these steps. The number of gene hits refers to how many gene targets were amplified during RT-qPCR across the triplicate samples: the qPCR method for the SDS samples targeted 3 genes for a total of 9 possible genes amplified while the method for the VTM samples targeted 2 genes for a total of 6 possible gene hits.
Study findings
All the cleaning methods performed well at low viral load over five cleaning cycles, and most cleaning failures were observed with BZK. While at higher viral loads, a combination of RNase AWAY and SDS effectively removed the viral signal, at low, medium, and high (all three) viral loads, the most effective cleaning solution was RNAse Away. While wet wipes and BZK performed well with SDS at medium viral loads, only wet wipes performed well with VTM under these conditions, thus indicating that wet wipes perform well in the medium viral load condition compared to BZK wipes.
As observed in past studies, SDS returned better signals than VTM on the same samples. Although repeat cleaning did not degrade the sentinel card surface or its ability to detect a viral signal, the authors recommended that at higher viral loads, replacement of the sentinel card was more effective than cleaning.
Conclusions
The study findings indicated that sentinel cards are an effective tool for SARS-CoV-2 classroom monitoring. However, they must be cleaned thoroughly at lower viral loads to remove carryover signal, and this process is more straightforward with samples collected in SDS than in VTM.
As cleaning the high viral load from sentinel cards could be more difficult, strong positives should be removed rather than cleaned. Overall, these findings support using the sentinel cards on a larger scale for SARS-CoV-2 monitoring in indoor settings.
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Sentinel Cards Provide Practical SARS-CoV-2 Monitoring in School Settings Victor J Cantú, Karenina Sanders, Pedro Belda-Ferre, Rodolfo A Salido, Rebecca Tsai, Brett Austin, William Jordan, Menka Asudani, Amanda Walster, Celestine G. Magallanes, Holly Valentine, Araz Manjoonian, Carrissa Wijaya, Vinton Omaleki, Stefan Aigner, Nathan A Baer, Maryann Betty, Anelizze Castro-Martínez, Willi Cheung, Peter De Hoff, Emily Eisner, Abbas Hakim, Alma L Lastrella, Elijah S Lawrence, Toan T Ngo, Tyler Ostrander, Ashley Plascencia, Shashank Sathe, Elizabeth W Smoot, Aaron F Carlin, Gene W Yeo, Louise C Laurent, Anna Liza Manlutac, Rebecca Fielding-Miller, Rob Knight, bioRxiv, 2022.02.01.478759; doi: https://doi.org/10.1101/2022.02.01.478759, https://www.biorxiv.org/content/10.1101/2022.02.01.478759v1
Posted in: Child Health News | Device / Technology News | Medical Research News | Disease/Infection News
Tags: Coronavirus, Coronavirus Disease COVID-19, Digital PCR, Education, Gene, Genes, Genomic, heat, Labor, Respiratory, RNA, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Virus

Written by
Neha Mathur
Neha Mathur has a Master’s degree in Biotechnology and extensive experience in digital marketing. She is passionate about reading and music. When she is not working, Neha likes to cook and travel.
Source: Read Full Article