New single-cell prenatal blood test can identify genetic abnormalities

Non-invasive prenatal tests (NIPTs) are used for fetal genetic disease screening in pregnant women. In contrast, invasive tests like amniocentesis carry the risk of causing fetal harm. A report in The Journal of Molecular Diagnostics describes the development of a single-cell DNA assessment method with high sensitivity and specificity. This noninvasive test enables direct extraction of genetic information from live fetal cells for simultaneous evaluation, thereby improving the likelihood of detecting a fetal anomaly.
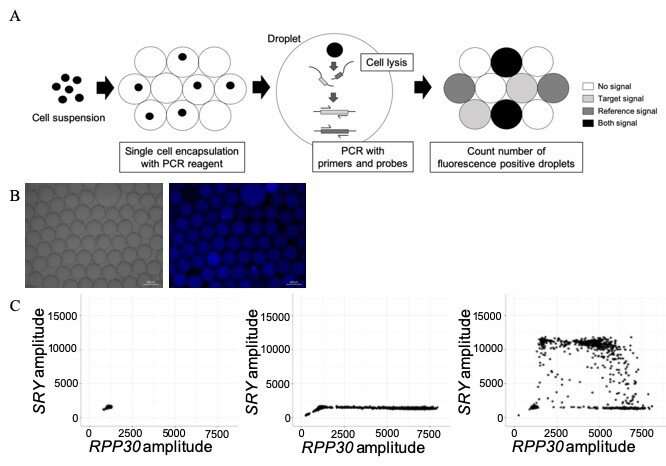
Established NIPTs have important limitations including noninformative results. Therefore, novel methods for noninvasive definitive diagnosis of fetal genetic abnormalities are needed. Using a modified single-cell-based droplet digital PCR (sc-ddPCR) NIPT, researchers conducted a proof of concept study that successfully assessed the genetic information of extremely rare fetal cells in maternal peripheral blood. This modified sc-ddPCR system makes it possible to directly assess single-cell DNA information from live cells without cell-fixation, cell-staining, and whole genome amplification steps.
“Because NIPTs currently analyze small DNA fragments, it can be challenging to ascertain whether the origin of each DNA fragment is the mother or her fetus,” explained lead investigator Kenichiro Hata, MD, Ph.D., Chairman of Maternal-Fetal Biology at the National Research Institute for Child Health and Development, Tokyo, Japan. “We have observed that in some cases NIPT results are discordant with the fetal genetic information that has been reported. This study serves as a proof of concept for noninvasive prenatal diagnosis using circulating fetal cells without any strict cell purification.”
With this modified technique, up to 3,000 cells per well can be encapsulated in each droplet and simultaneously analyzed. The original sc-ddPCR system simultaneously assesses single-cell genetic information with high sensitivity and specificity. However, the original system is only useful for cell suspensions with a clear background (i.e., washed cell lines or clearly sorted cells with fluorescence-activated cell sorting). Researchers modified this sc-ddPCR system to improve the PCR environment in each droplet with higher sensitivity and specificity, which makes it possible to now assess single-cell genetic information from crudely purified nucleated cell samples with impurities.
To confirm the sensitivity of this modified sc-ddPCR system, investigators detected the genomic DNA of circulating male fetal cells in a crudely sorted cell suspension at the single-cell level derived from peripheral blood samples from mothers with male fetuses.
Investigators searched for the presence of the sex-determining region Y gene (SRY), which is responsible for the initiation of male sex determination. Analysis of 13 blood samples indicated that only circulating fetal cells from the three pregnant women carrying male fetuses tested positive for the SRY gene, unlike cells from the 10 pregnant women carrying female fetuses. This indicates that the modified sc-ddPCR system not only has high sensitivity, but also high specificity.
Source: Read Full Article