Visualization of SARS-CoV-2 RNA by fluorescence in situ hybridization probes

A team of scientists from France has recently designed fluorescence in situ hybridization probes (CoronaFISH) to visualize severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA in infected cells. A detailed description of the probe design, validation, and application is currently available on the bioRxiv* preprint server.
Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative pathogen of coronavirus disease 2019 (COVID-19), is an enveloped, positive-sense, single-stranded RNA virus belonging to the family Coronaviridae. The single linear RNA segment of SARS-CoV-2 serves as a template for viral transcription and replication. The RNA-dependent RNA polymerase of the virus generates a negative-sense strand, which is used as a template for replicating full-length viral RNA genomes and transcription of subgenomic positive-stranded RNAs (sgRNAs). Subsequently, viral proteins are synthesized from these sgRNAs and mature virions are released from the infected cells via exocytosis.
To explore the subcellular localization and kinetics of RNA viruses in infected cells, it is important to directly and precisely visualize the viral RNA in single cells. In the current study, the scientists have designed and validated single-molecule CoronaFISH probes against the positive and negative RNA strands of SARS-CoV-2. They have used these probes for visualizing viral RNA in cells isolated from African green monkey, human cell lines, COVID-19 patient samples, and human tissues.
Important observations
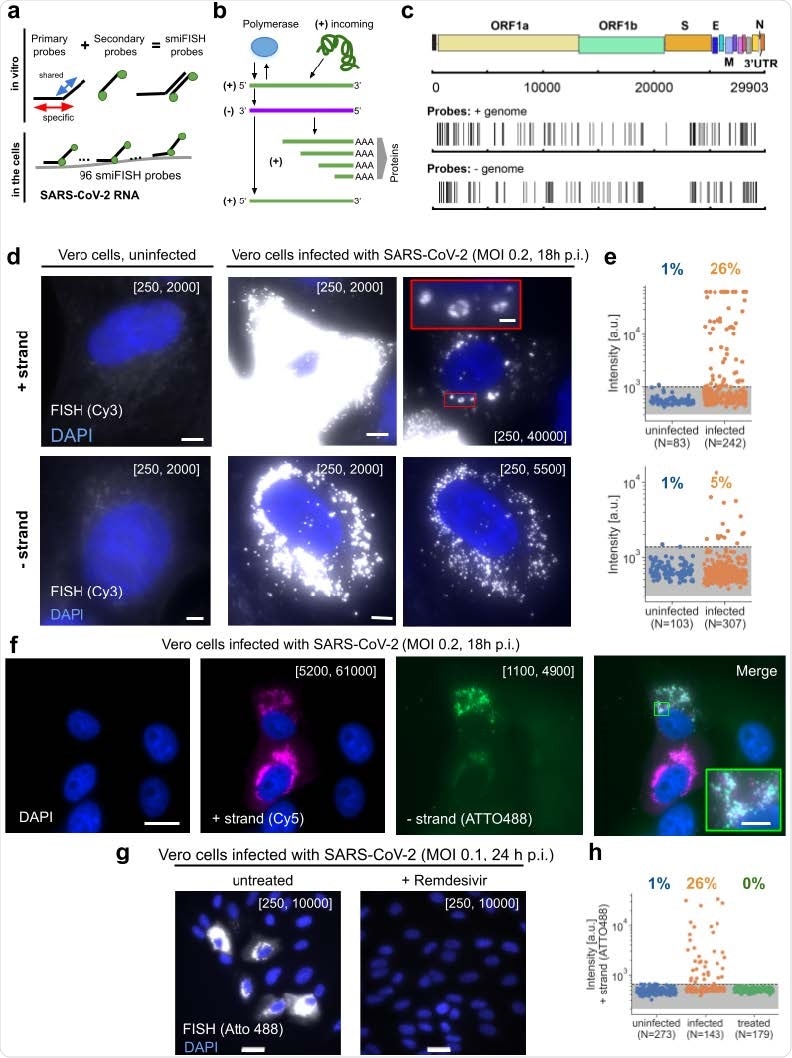
To visualize SARS-CoV-2 RNA in infected cells, the scientists used positive and negative-stranded probes labeled with fluorophore Cy3. They observed only weak and diffuse fluorescent signal in uninfected cells, whereas in infected cells, they observed very strong and localized signal in the cytoplasm. The strong RNA signal observed in the cytoplasm supports the cytoplasmic replication cycle of SARS-CoV-2. The scientists believe that because the probes are designed to cover the entire length of viral RNA (about 30 kb), the intensity of the signal should not be affected by viral mutations or partial RNA degradation.
The quantitative analysis of the signal intensity revealed that positive-strand probes generated significantly higher intensity signals than negative-strand probes, justifying a lower amount of negative-stranded RNA replication intermediates in infected cells.
Using dual-color imaging, the scientists demonstrated that these FISH probes can visualize both positive-stranded and negative-stranded viral RNAs simultaneously. For this experiment, they labeled the probes with two different fluorophores (Atto488 and Cy5). The images they obtained using these probes clearly showed positive- and negative-stranded RNAs in the same infected cells and in the same subcellular locations. The process of simultaneous visualization of both positive and negative RNAs can be potentially used to study intracellular dynamics of viral transcription and replication.
The experiments conducted using nasal swabs from COVID-19 patients revealed that probes specific for positive-stranded viral RNA produced a strong signal in a subset of cells. Moreover, a separate set of experiments revealed that FISH probes could detect viral RNA in human cell lines and human tissue samples collected from COVID-19 patients.
Study significance
According to the scientists, the CoronaFISH probes they designed have several advantages over the immunofluorescence technique. Instead of marking viral proteins, these probes can directly visualize the viral genome, which is beneficial in terms of specifically examining the presence of the virus and its replication process in specific subcellular locations. In this way, it is possible to differentiate between primary SARS-CoV-2 infection (productive infection) and treatment-induced secondary infection (non-productive infection).
CoronaFISH can provide in-depth information about the presence and abundance of viral RNA in infected cells, subcellular localization of viral RNA, and host–virus interaction dynamics at the molecular level. Moreover, the probes can be modified for electron microscopic visualization of SARS-CoV-2 RNA.
Another advantage of CoronaFISH is unique complementary sequences between the probes and the virus, making it possible to distinguish SARS-CoV-2 from other viral strains. Moreover, the probes can be prepared rapidly and cost-effectively, which makes CoronaFISH an attractive choice for high-throughput image-based screening of therapeutic compounds.
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Sensitive visualization of SARS-CoV-2 RNA with CoronaFISH Elena Rensen, Stefano Pietropaoli, Christian Weber, Sylvie Souquere, Pierre Isnard, Marion Rabant, Jean-Baptiste Gibier, Etienne Simon-Loriere, Marie-Anne Rameix-Welti, Gérard Pierron, Florian Mueller, Giovanna Barba-Spaeth, Christophe Zimmer, bioRxiv, 2021.02.04.429604; doi: https://doi.org/10.1101/2021.02.04.429604, https://www.biorxiv.org/content/10.1101/2021.02.04.429604v1
Posted in: Device / Technology News | Medical Research News | Disease/Infection News
Tags: Cell, Coronavirus, Coronavirus Disease COVID-19, Cytoplasm, Electron, Fish, Fluorescence, Fluorophore, Genome, Genomic, Hybridization, Imaging, in vitro, Intracellular, Molecule, Pathogen, Polymerase, Quantitative Analysis, Remdesivir, Respiratory, RNA, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Transcription, Virus

Written by
Dr. Sanchari Sinha Dutta
Dr. Sanchari Sinha Dutta is a science communicator who believes in spreading the power of science in every corner of the world. She has a Bachelor of Science (B.Sc.) degree and a Master's of Science (M.Sc.) in biology and human physiology. Following her Master's degree, Sanchari went on to study a Ph.D. in human physiology. She has authored more than 10 original research articles, all of which have been published in world renowned international journals.
Source: Read Full Article