Precision insights can be found in wastewater


Research from the lab of Fangqiong Ling at Washington University in St. Louis showed earlier this year that the amount of SARS-CoV-2 in a wastewater system was correlated with the burden of disease—COVID-19—in the region it served.
But before that work could be done, Ling needed to know: How can you figure out the number of individuals represented in a random sample of wastewater?
A chance encounter with a colleague helped Ling, an assistant professor in the Department of Energy, Environmental and Chemical Engineering at the McKelvey School of Engineering, develop a machine learning model that used the assortment of microbes found in wastewater to tease out how many individual people they represented. Going forward, this method may be able to link other properties in wastewater to individual-level data.
The research was published in the journal PLOS Computational Biology.
The problem was straightforward: “If you just take one scoop of wastewater, you don’t know how many people you’re measuring,” Ling said. This is counter to the way studies are typically designed.
“Usually when you design your experiment, you design your sample size, you know how many people you’re measuring,” Ling said. Before she could look for a correlation between SARS-CoV-2 and the number of people with COVID, she had to figure out how many people were represented in the water she was testing.
Initially, Ling thought that machine learning might be able to uncover a straightforward relationship between the diversity of microbes and the number of people it represented, but the simulations, done with an “off-the-shelf” machine learning, didn’t pan out.
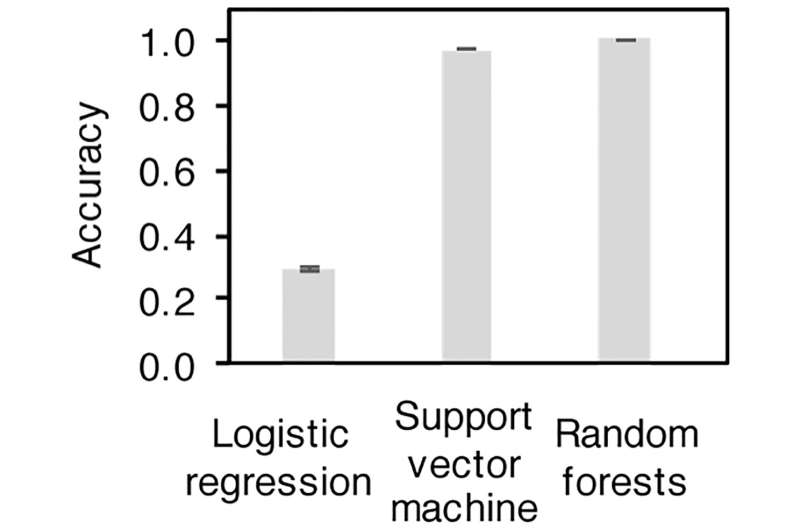
Then Ling had a chance encounter with Likai Chen, an assistant professor of mathematics and statistics in Arts & Sciences. The two realized they shared an interest in working with novel, complex data. Ling mentioned that she was working on a project that Chen might be able to contribute to.
“She shared the problem with me and I said, that’s indeed something we can do,” Chen said. It happened that Chen was working on a problem that used a technique that Ling also found helpful.
The key to being able to tease out how many individual people were represented in a sample is related to the fact that, the bigger the sample, the more likely it is to resemble the mean, or average. But in reality, individuals tend not to be exactly “average.” Therefore, if a sample looks like an average sample of microbiota, it’s likely to be made up of many people. The farther away from the average, the more likely it is to represent an individual.
“But now we are dealing with high-dimensional data, right?” Chen said. There are near-endless number of ways that you can group these different microbes to form a sample. “So that means we have to find out, how do we aggregate that information across different locations?”
Using this basic intuition—and a lot of math—Chen worked with Ling to develop a more tailored machine learning algorithm that could, if trained on real samples of microbiota from more than 1,100 people, determine how many people were represented in a wastewater sample (these samples were unrelated to the training data).
“It’s much faster and it can be trained on a laptop,” Ling said. And it’s not only useful for the microbiome, but also, with sufficient examples—training data—this algorithm could use viruses from the human virome or metabolic chemicals to link individuals to wastewater samples.
“This method was used to test our ability to measure population size,” Ling said. But it goes much further. “Now we are developing a framework to allow validation across studies.”
More information:
Lin Zhang et al, MicrobiomeCensus estimates human population sizes from wastewater samples based on inter-individual variability in gut microbiomes, PLOS Computational Biology (2022). DOI: 10.1371/journal.pcbi.1010472
Journal information:
PLoS Computational Biology
Source: Read Full Article