Novel newborn screening system uses rapid whole genome sequencing to screen and diagnose genetic diseases

Rady Children’s Institute for Genomic Medicine (RCIGM) today announced the publication of a study in the American Journal of Human Genetics (AJHG) describing a scalable prototype for newborn screening (NBS) that includes the diagnosis of genetic diseases by rapid Whole Genome Sequencing (rWGS) and virtual acute management guidance.
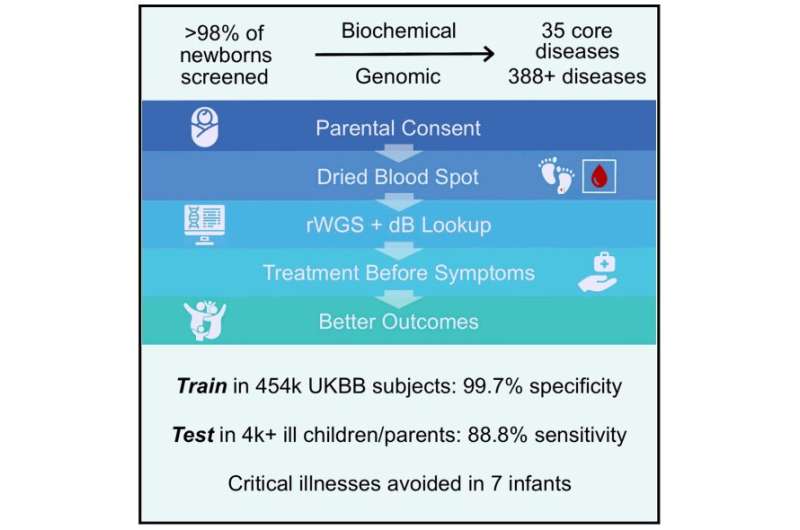
“This NBS-rWGS system is designed to complement the existing newborn screening process and has the potential to eliminate the diagnostic and therapeutic odyssey that many children and parents face,” said Stephen Kingsmore, MD, DSc, President & CEO of RCIGM and lead author. “Currently, only 35 core genetic disorders are recommended for newborn screening in the United States, but there are more than 7,200 known genetic diseases. Outcomes remain poor for newborns with a genetic disease because of the limited number of recommended screenings. With NBS-rWGS, we can more quickly expand that number and therefore potentially improve outcomes through precision medicine.”
In the study, the authors used established criteria, a Delphi technique, electronic data capture, and consensus from a panel of pediatric geneticists on six questions to determine inclusion of interventions and disorders in Genome-to-Treatment (GTRx), an automated, virtual disease management system that integrates a rWGS diagnosis with a custom lab information management system and analysis pipeline. They reviewed 457 genetic diseases and selected 388 for inclusion in NBS-rWGS. After simulating NBS-rWGS in 454,707 UK Biobank subjects with 29,865 pathogenic or likely pathogenic variants associated with these 388 genetic diseases, they determined that NBS-rWGS had specificity of 99.7%.
In 2,208 critically ill children with suspected genetic disorders, and 2,168 of their parents, the researchers found that simulated NBS-rWGS identified 104 of 119 diagnoses previously made by rWGS, and identified 15 findings not previously reported. The negative predictive value and sensitivity of NBS-rWGS (99.6% and 88.8%, respectively) were slightly higher than diagnostic rWGS. Earlier interventions may have markedly improved outcomes and reduced hospitalization in 43 of the 104 critically ill children.
The simulated NBS-rWGS identified genetic diseases in 15 newborns that had been missed by conventional newborn screening. In addition, in 60 of 104 children with genetic diseases identified by simulated NBS-rWGS, the researchers calculated that performing NBS-rWGS within five days of birth may have shortened the time to diagnosis by a median of 73 days. They determined that this could have potentially avoided symptoms completely in seven of those newborns.
“The results of this study demonstrate the potential for newborn screening by rapid Whole Genome Sequencing to help shorten the time to diagnosis for hundreds of genetic diseases,” said Tom Defay, co-author and Alexion’s Deputy Head, Diagnostics Strategy and Development. “As part of our commitment to help patients and their families get answers faster, we are continuing to collaborate with Rady Children’s Institute for Genomic Medicine as they work to enhance and scale this innovative technology.”
Source: Read Full Article