Wastewater monitoring reveals emergence and spread of SARS-CoV-2 Omicron in Alberta

The onset of the coronavirus disease 2019 (COVID-19) pandemic led to scientific progress in wastewater-based surveillance of community infections. Moreover, the measurement of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) ribonucleic acid (RNA) in sewage samples was used as a complementary surveillance tool during the early stages of the pandemic.

Study: Emergence and Spread of the SARS-CoV-2 Omicron Variant in Alberta Communities Revealed by Wastewater Monitoring. Image Credit: boonchoke / Shutterstock.com
Soon after, hundreds of wastewater COVID-19 monitoring groups and online dashboards were established throughout the world, including in Alberta. This strategy was based on the fecal shedding of individuals infected with SARS-CoV-2 who could be diagnosed by quantification of the viral RNA in sewage sampled at wastewater treatment plants (WWTPs) or other nodes.
Research teams located in Alberta and other places around the world have suggested that wastewater is a leading indicator of COVID-19 during the pandemic waves since these results have preceded clinically diagnosed cases by four to six days. Thus, sampling, testing, and reporting of viral RNA in wastewater can provide early warning of the population-wide disease burden that, in turn, can help with decision-making processes on disease control.
Background
South Africa first detected the SARS-CoV-2 Omicron variant in the Gauteng province on November 24, 2021. The Omicron variant was quickly labeled as a SARS-CoV-2 variant of concern (VOC) by the World Health Organization on November 26, 2021.
Soon after, the Omicron variant was rapidly detected throughout the world. By January 2022, the Omicron variant had become the dominant circulating strain in most countries around the world, thus prompting the re-introduction of public health restrictions.
Studies have suggested that wastewater testing could differentiate changes in the disease burden caused by different VOCs in communities. Sequencing of viral genomes from wastewater can be achieved through targeted amplicon tiling protocols or shotgun metagenomics; however, these approaches are time-consuming and costly. Comparatively, targeted reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assay of the RNA from wastewater has provided accurate data on VOCs at a lower cost and in near-real-time.
In a new study published on the preprint server medRxiv,* researchers utilize variant-specific RT-qPCR assays to monitor wastewater in 30 municipalities up to three times per week to determine the emergence and temporal change in the prevalence of the Omicron and Delta variants in Alberta.
About the study
The current study involved the collection of wastewater samples from municipal wastewater treatment plants (WWTPs) up to three times per week. RNA was then isolated from the wastewater either by affinity binding columns or ultrafiltration, followed by RNA extraction. Notably, wastewater collected from Calgary, Lethbridge, and Fort McMurray was processed using both methods.
The quantification of RNA was achieved through RT-qPCR. Delta, Omicron, and total SARS-CoV-2 assays were triplexed together to estimate an Omicron-to-Delta ratio in each wastewater sample. Additionally, the daily number of new clinically diagnosed COVID-19 cases were collected from Data Analytics of Alberta Health Services.
Study findings
The Omicron variant was first detected in the wastewater of the Alberta community during late November and early December. In fact, between 3% and 9% of samples collected between December 5, 2021, to December 9, 2021, exhibited a sustained presence of Omicron in Calgary. The detection of Omicron in the capital city of Edmonton was reported on December 10, 2021.
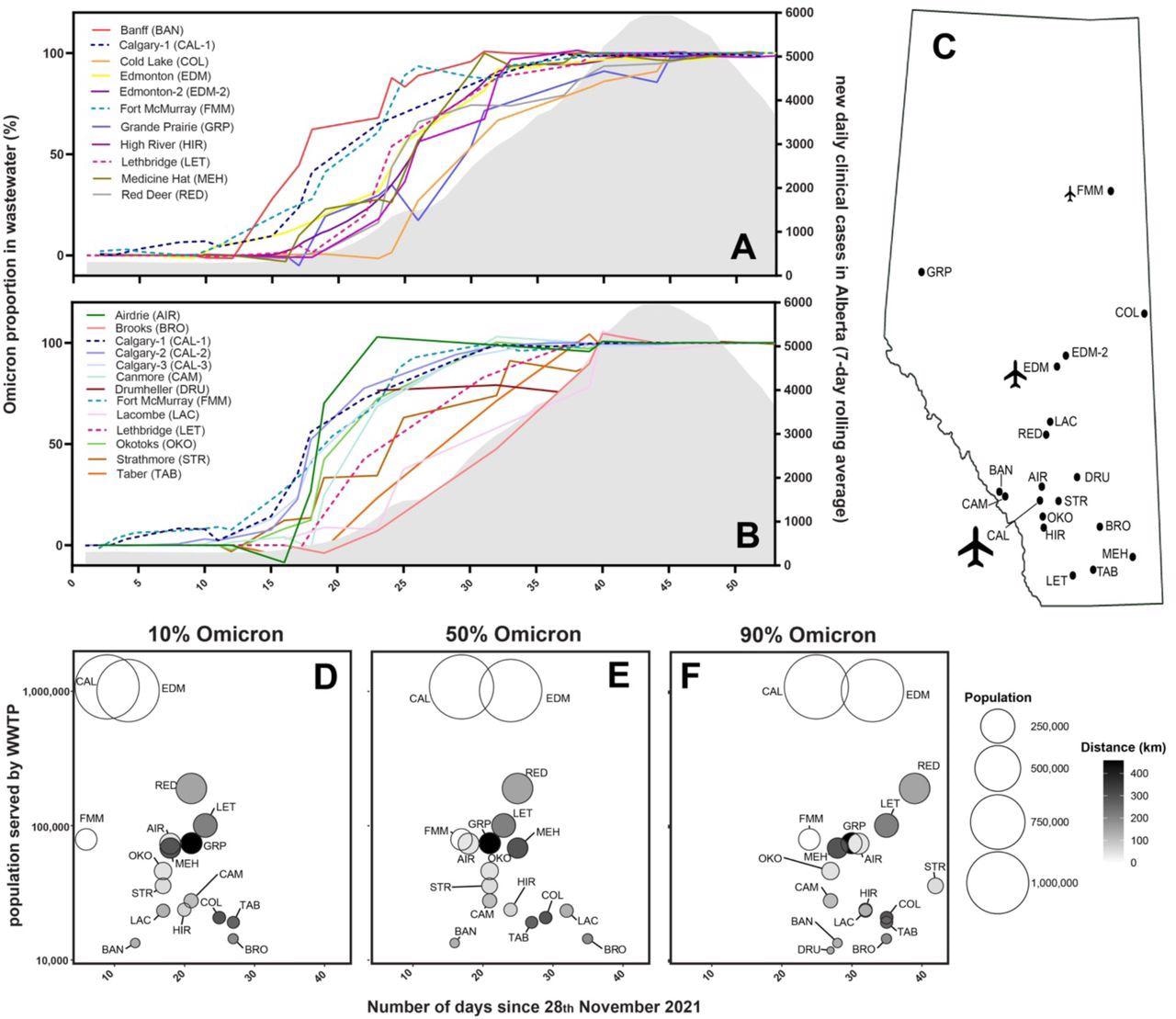
Proportion of Omicron relative to Delta SARS-CoV-2 variants in community wastewater samples assessed using RT-qPCR assays for specific variants following sample processing using ultrafiltration (A) or affinity columns (B). Lines of best fit plotted with second order smoothing are shown for different WWTPs, including 3 that had samples processed using both ultrafiltration (A) and affinity columns (B) for comparison (see dashed lines for Calgary-1, Fort McMurray and Lethbridge; Fig. S1). Monitoring lasted for 53 days beginning on November 28th (plotted as consecutive days on the x-axes). The grey shaded area on the right side (A, B) shows the 7-day rolling average of new clinical cases reported in Alberta (right y-axis), which increased after the Omicron variant was predominant in municipal wastewater from 30 communities sampled at 21 WWTPs throughout the province (C). Calgary and Edmonton are served by 3 and 2 WWTPs, respectively (A, B), and some individual WWTPs also serve several municipalities (e.g., Edmonton-2 serves 6 others; Red Deer serves 3 others; Calgary’s WWTPs serve 3 others). The timing (in days) of the Omicron-to-Delta ratio passing 10%, 50% and 90% of community COVID-19 burden (D-F) reveals general trends of decreasing population size (bubble diameter and y-axis) and distance from the nearest airport in Calgary, Edmonton or Fort McMurray (bubble shading). Bubble plots only include data from Calgary-1 and Edmonton-1 WWTPs (the largest WWTP from each city), scaled to the population of the corresponding sewershed sub-catchment in those cities.
The rate of increase of Omicron in the international resort town of Banff was higher as compared to larger cities such as Edmonton and Calgary. Omicron was found to have surpassed 80% of all community cases in Banff, while it had surpassed just 50% of new cases in Calgary and Edmonton.
The proportion of Omicron infections was also found to grow in smaller bedroom communities that were located adjacent to Edmonton and Calgary. Smaller and remote communities such as Brooks experienced the most delayed emergence of Omicron and did not reach higher proportions until December 29, 2021.
Conclusions
The current study demonstrates that wastewater testing can provide an unbiased and accurate representation of disease prevalence, especially during the early stages of the disease. This approach is also cost-effective and can help provide objective information to policymakers and public-health authorities in near-real-time. Therefore, wastewater testing can be adapted by other large jurisdictions for the detection of emerging pathogens.
*Important notice
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Hubert, C. R. J., Acosta, N., Waddell, B. J., et al. (2022). Emergence and Spread of the SARS-CoV-2 Omicron Variant in Alberta Communities Revealed by Wastewater Monitoring. medRxiv. doi:10.1101/2022.03.07.22272055. https://www.medrxiv.org/content/10.1101/2022.03.07.22272055v1.
Posted in: Medical Research News | Disease/Infection News
Tags: Assay, Coronavirus, Coronavirus Disease COVID-19, covid-19, Diagnostics, Metagenomics, Omicron, Pandemic, Polymerase, Polymerase Chain Reaction, Public Health, Research, Respiratory, Ribonucleic Acid, RNA, RNA Extraction, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Transcription

Written by
Suchandrima Bhowmik
Suchandrima has a Bachelor of Science (B.Sc.) degree in Microbiology and a Master of Science (M.Sc.) degree in Microbiology from the University of Calcutta, India. The study of health and diseases was always very important to her. In addition to Microbiology, she also gained extensive knowledge in Biochemistry, Immunology, Medical Microbiology, Metabolism, and Biotechnology as part of her master's degree.
Source: Read Full Article