Proximity labeling to identify high-confidence SARS-CoV-2 interactors

A recent report published in the STAR Protocols journal depicted a protocol for an antibody-based proximity labeling to discover biotinylated interactors of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
Study: An antibody-based proximity labeling protocol to identify biotinylated interactors of SARS-CoV-2. Image Credit: Star Protocols
Background
The interplay between host and SARS-CoV-2 components is critical for the viral life cycle in coronavirus disease 2019 (COVID-19). The molecular mechanisms of viral pathogenicity are poorly understood due to the complexity of the virus-host interaction. Understanding viral protein activities during infection require depicting the virus-host interactome.
A streptavidin enrichment approach was recently used to examine proximal proteins of SARS-CoV-2 based on BioID, a technique for identifying protein interactions in live cells. Nevertheless, the biotinylated peptides were hard to pinpoint because of the significant binding affinity of biotin and streptavidin, which was crucial for measuring the certainty of proximity contacts.
About the study
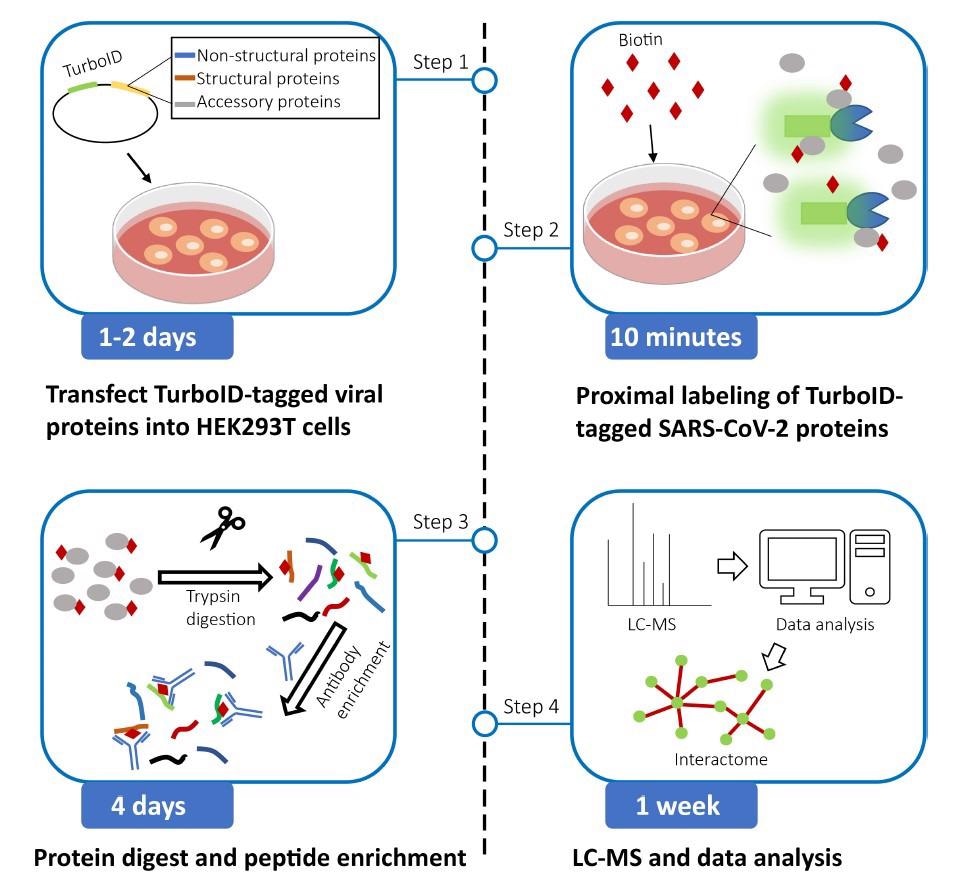
In the present study, the authors provided a protocol to explore SARS-CoV-2 protein interactors employing the TurboID proximity labeling technique based on an antibody. TurboID is a biotin ligase that converts biotin to biotin–adenosine monophosphate (AMP) using adenosine triphosphate (ATP). The scientists utilized a biotin-specific antibody to enhance proximal proteins of SARS-CoV-2 with biotinylated peptides labeled with TurboID-tagged viral proteins.
The team explained the processes for preparing biotinylated peptide samples for mass spectrometry (MS) analysis. They also described a strict workflow for identifying biotinylated high-confident interactors of SARS-CoV-2 via screening out unspecific co-purified proteins.
Protocol in-detail
The authors constructed the expression vectors for SARS-CoV-2 proteins over six weeks. TurboID expression vectors with SARS-CoV-2 protein-coding sequences were used to generate TurboID-tagged viral proteins, and their expression was verified. The authors stated that the transfection reagent could be substituted with Turbofect or Lipofectamine 2000.
Proximity labeling by TurboID-tagged SARS-CoV-2 proteins was carried out over a week. TurboID-tagged SARS-CoV-2 expression vectors were transfected into human embryonic kidney 293T (HEK293T) cells to biotinylate proximal proteins. The researchers highlighted the need for extreme caution while changing the culture media to avoid cell loss since HEK293T cells were easily severed from plates. The sonication's pulse time and power capacity could be adjusted till the achievement of clear cell lysis. The streptavidin-Horseradish peroxidase (HRP) and bovine serum albumin (BSA) blocking buffer should be freshly prepared.
Protein digestion by filter aided sample preparation (FASP) was conducted over two days. Cell lysis proteins were alkylated, minimized, and digested in centrifugal filter units. The researchers procured and dried the digested peptide combinations.
Likewise, peptide enhancement and desalting were also conducted in two days. Digested peptides were augmented using the anti-biotin antibody. StageTips were used to desalt the extracted peptides, and vacuum centrifugation was used to dry them. Finally, the team performed MS and data analysis over a week.
Anticipated results
The authors mentioned that the biotinylated peptides labeled by the SARS-CoV-2 proteins tagged by TurboID were directly identified using the present approach. After rigorous data processing, these significantly enhanced proteins with biotinylated regions in sample groups were identified as high-confidence interactors of SARS-CoV-2 proteins.
The generation of TurboID-tagged SARS-CoV-2 proteins and their capacities to mark proximal interactors allows a thorough interactome investigation of host components and viral proteins. The present protocol employed an anti-biotin antibody to acquire proximal proteins harboring biotinylated sites. This resulted in the recognition of 1388 high-confidence proximal interacting molecules of SARS-CoV-2 proteins, of which 1092 were not explored by the streptavidin-based BioID analysis in the SARS-CoV-2 interactome research. Thus, demonstrating the benefits of the antibody-based TurboID technique in pinpointing proximal interactors. The resultant data will aid in discovering SARS-CoV-2 pathogenesis and COVID-19 therapy development.
Resolution of problems
The authors suggested solutions for some of the problems that occurred during the analyses are as follows:
1. Poor transfection potential of TurboID-fusion expression vectors. Possible solution: The aliquoted polyetherimide (PEI) kept at -20℃ should be dissolved entirely at room temperature (RT). Insoluble particles could be heated at 65°C for almost half an hour till the solution becomes clear. In addition, minimize freeze-thaw cycles of PEI.
2. Western blots did not identify the expression of TurboID-tagged SARS-CoV-2 proteins. Possible solution: Substitution of cytomegalovirus (CMV) promoter with CMV enhancer, chicken β-actin promoter, and rabbit β-globin splice acceptor site (CAG) of the vector enhances their expression. For the eukaryotic expression vector, improve viral open reading frame (ORF) complementary deoxyribonucleic acid (cDNA) codons. Additionally, viral proteins and TurboID should be oriented alternatively.
3. Low expression of biotinylated proteins identified by Western blot. Possible solution: Harvest cells 48 hours after transfection for high-molecular-weight viral proteins leading to biotinylation and detection of proximal proteins.
4. Ceratin biotinylated proteins were drastically weak to be detected by Western blot using streptavidin-HRP. Possible solution: Certain TurboID-tagged viral proteins possess weaker biotinylation signals and engage with few proximal proteins than others. Always stack TurboID without and with biotin controls containing samples on the same membrane to validate sample biotinylation.
5. Biotinylated proteins were recognized effectively, although liquid chromatography with tandem MS (LC-MS-MS) only identified fewer proteins. Possible solution: This might be attributed to protein loss in the bead washing process. Do not pipette up and down a lot to avoid beads sticking to the pipette tips. Aspirate the supernatant carefully without beads.
- Limin Shang, Yuehui Zhang, Yuchen Liu, Chaozhi Jin, Yanan Zhao, Jing Zhang, Pei-Hui Wang, Jian Wang, An antibody-based proximity labeling protocol to identify biotinylated interactors of SARS-CoV-2, STAR Protocols, 2022, 101406, ISSN 2666-1667. DOI: https://doi.org/10.1016/j.xpro.2022.101406, https://www.sciencedirect.com/science/article/pii/S2666166722002866
Posted in: Molecular & Structural Biology | Medical Science News | Disease/Infection News
Tags: Actin, Adenosine, Adenosine Triphosphate, Albumin, Antibody, binding affinity, Bovine Serum Albumin, Cell, Cell Lysis, Chromatography, Coronavirus, Coronavirus Disease COVID-19, covid-19, Cytomegalovirus, Digestion, Interactome, Kidney, Ligase, Liquid Chromatography, Mass Spectrometry, Membrane, Peptides, Promoter, Protein, Research, Respiratory, Sample Preparation, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Spectrometry, Syndrome, Transfection, Virus, Western Blot

Written by
Shanet Susan Alex
Shanet Susan Alex, a medical writer, based in Kerala, India, is a Doctor of Pharmacy graduate from Kerala University of Health Sciences. Her academic background is in clinical pharmacy and research, and she is passionate about medical writing. Shanet has published papers in the International Journal of Medical Science and Current Research (IJMSCR), the International Journal of Pharmacy (IJP), and the International Journal of Medical Science and Applied Research (IJMSAR). Apart from work, she enjoys listening to music and watching movies.
Source: Read Full Article