Human Body-on-Chip platform enables in vitro prediction of drug behaviors in humans

Drug development is an extremely arduous and costly process, and failure rates in clinical trials that test new drugs for their safety and efficacy in humans remain very high. According to current estimates, only 13.8% of all tested drugs demonstrate ultimate clinical success and obtain approval by the Food and Drug Administration (FDA). There are also increasing ethical concerns relating to the use of animal studies. As a result, there has been a world-wide search to find replacements for animal models.
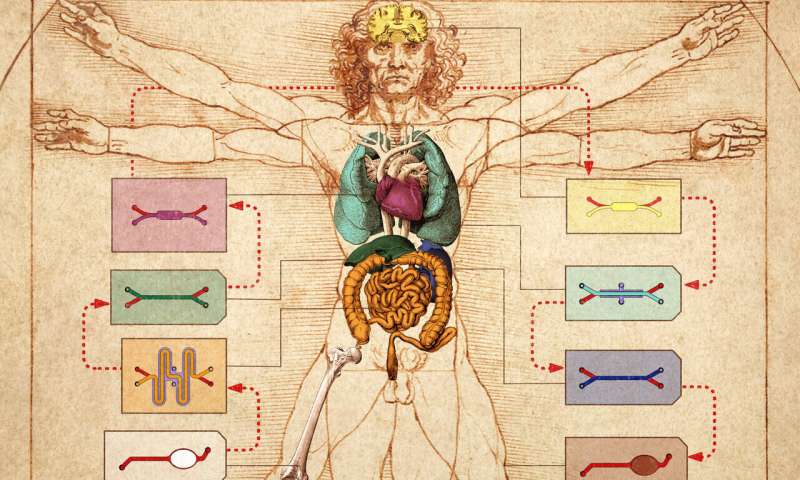
To help address this bottleneck in drug development, Donald Ingber, M.D., Ph.D., and his team at Harvard’s Wyss Institute for Biologically Inspired Engineering, developed the first human “Organ-on-a-Chip” (Organ Chip) model of the lung that recapitulates human organ level physiology and pathophysiology with high fidelity, which was reported in Science in 2010. Organ Chips are microfluidic culture devices composed of a clear flexible polymer the size of a computer memory stick, which contains two parallel hollow channels that are separated by a porous membrane. Organ-specific cells are cultured on one side of the membrane in one of the channels, and vascular endothelial cells recapitulating a blood vessel line the other, while each channel is independently perfused with cell type-specific medium. The porous membrane allows the two compartments to communicate with each other, and to exchange molecules like cytokines, growth factors, and drugs, as well as drug breakdown products generated by organ-specific metabolic activities.
One example where living animals must be used in preclinical testing is the characterization of a drug’s “pharmacokinetics” (PK) that involves the quantification of its absorption, distribution, metabolism, and excretion (ADME), which together determine drug levels in the blood. These responses involve interplay between many different organs linked by a vasculature containing flowing blood. Animals are also used to analyze drug “pharmacodynamics” (PD), the effects the drug produces on its target organs, which underlies its mechanism of action as well as its adverse effects.
Because the Wyss Institute’s Organ Chips contain an endothelium-lined vascular channel, Ingber proposed in 2011 that it might be possible to create a human “Body-on-Chips” by transferring fluids between the vascular channels of many different types of Organ Chips to mimic blood flow, and assessing drug PK/PD behaviors across the entire linked system. Inspired by this vision and the realization that existing animal-based development programs are inadequate to confront the needs for accelerated development of drug countermeasures in a biothreat situation, the Defense Advanced Research Projects Agency (DARPA) requested grant applications in 2012 with a seemingly impossible challenge: develop 10 types of Organ Chips that recapitulate the complex functionalities of 10 different human organs, engineer an automated instrument to fluidically link them to create a functional human Body-on-Chips platform, and leverage computational modeling in combination with experimental data generated using this platform to quantitatively predict human drug PK/PD behavior in vitro. Now, two back-to-back publications in Nature Biomedical Engineering, describe the Wyss team’s success in meeting this goal in full.
Known for posing impossible challenges such as this, DARPA understands that most investigators will not meet the goals as set out, but that extraordinary technological fallout will be created along the way. “We were very proud to obtain major funding support from DARPA to take on this challenge, and we are now even more proud that we have successfully met their goal, which would not have been possible without the exceptional talents, interdisciplinary spirit, and monumental team effort at the Wyss Institute,” said Ingber, who is the Wyss Founding Director, as well as the Judah Folkman Professor of Vascular Biology at Harvard Medical School and Boston Children’s Hospital, and Professor of Bioengineering at the Harvard John A. Paulson School of Engineering and Applied Sciences (SEAS). He has been leading the DARPA-funded program along with Wyss Core Faculty member Kevin Kit Parker, Ph.D., who also is the Tarr Family Professor of Bioengineering and Applied Physics at SEAS.
In their first article, the Wyss team presents a highly modular Body-on-Chips platform, which is enabled by an engineered “Interrogator” instrument that can culture up to 10 different Organ Chips, and sequentially transfer fluids between their endothelium-lined vascular channels to mimic normal human blood flow between the different organs of our body. In the second article, the team uses a computational scaling method to translate data obtained from drug experiments involving 3 different types of fluidically-linked Organ Chips to their respective organ dimensions in the real human body. The approach is able to quantitatively predict changes in drug levels over time, as well as organ-specific toxicities, that have been previously measured in human patients.
“Both studies represent a tremendous effort by scores of researchers at the Wyss Institute, who worked together with our industrial modeling collaborators, and pooled their collective tissue engineering, microfabrication, pharmacological, physiological, and computational expertise to make this huge advance in preclinical drug testing possible,” said Rachelle Prantil-Baun, Ph.D., a Wyss Institute Senior Staff Scientist with past experience in the pharmaceutical industry who helped to orchestrate this complex multi-investigator effort with multiple other staff members in the Wyss’ Bioinspired Therapeutics and Diagnostics platform.
The Interrogator instrument enabled the team to culture, perfuse and link many living human cultured tissues in a multi-Organ Chip system, as well as add and sample the medium in a fully programmable way, using the device’s robotic liquid transfer capabilities, while continuously monitoring tissue integrity with an integrated microscope. “In this study, we serially linked the vascular channels of eight different Organ Chips, including intestine, liver, kidney, heart, lung, skin, blood-brain barrier and brain, using a highly optimized common blood substitute, while independently perfusing the individual channels lined by organ-specific cells. The instrument maintained the viability of all tissues and their organ-specific functions for over three weeks and, importantly, it allowed us to quantitatively predict the tissue-specific distribution of a chemical across the entire system,” said Richard Novak, Ph.D., a co-first-author on both studies. Novak is a Senior Staff Engineer at the Wyss Institute who designed, fabricated, and operated the Interrogator instrument with his bioengineering team.
In the second study, the team used the Interrogator instrument to support two different configurations of 3 different Organ Chips linked to each other and to a central arterio-venous (AV) fluid mixing reservoir that helped recapitulate life-like blood and drug exchange between the individual organs, while also providing a way to carry out blood sampling that would mimic blood drawing from a peripheral vein. The researchers coupled a human Gut Chip with a Liver Chip and a Kidney Chip and added nicotine to the Gut Chip’s channel lined by intestinal epithelium to simulate oral administration of this drug, and its first pass through the intestinal wall and via the vascular system to the liver where it is metabolized, and to the kidney where it is excreted. Nicotine chewing gum is used to help with smoking cessation; however, it is also being investigated as an oral drug for neurodegenerative and inflammatory bowel diseases.
Applying mass spectrometry analysis, the Wyss team quantified nicotine levels in the AV reservoir and the effluents of the vascular channels of all the different Organ Chips, and then fitted the data with a newly developed biomimetic scaling approach that translates them from the dimensions of the Organ Chips to their actual organ dimensions in the human body. For the first time, this computational approach combined with experimental human Organ Chip data demonstrated the ability to model drug uptake and metabolism, and quantitatively predict dynamic changes in drug blood levels (PK) that previously were observed in human clinical trials. The scaling approach, which also solves the challenge of drug adsorption into materials in the experimental system, was developed by co-author Andrzej Przekwas, Ph.D., and his team at CFD Research Corporation in Huntsville, Alabama.
“The resulting calculated maximum nicotine concentrations, the time needed for nicotine to reach the different tissue compartments, and the clearance rates in the Liver Chips in our in vitro-based in silico model mirrored closely what had been measured previously in patients,” said Ben Maoz, Ph.D., a co-first author on the second study and former Technology Development Fellow at the Wyss Institute in the lab of Parker. Maoz currently is an Assistant Professor at Tel Aviv University, Israel.
With a second multi-Organ Chip configuration comprising fluidically-linked Liver, Kidney, and Bone Marrow Chips, the team investigated the pharmacological effects of cisplatin, a chemotherapeutic drug commonly used in cancer treatments, which is administered intravenously and displays unwanted toxicity in the kidney and bone marrow. “Our analysis recapitulates the pharmacodynamic effects of cisplatin in patients, including a decrease in numbers of different blood cell types and an increase in markers of kidney injury,” said co-first author Anna Herland, Ph.D., who worked on Ingber’s team at the time of the study. “In addition, the in vitro-to-in vivo translation capabilities of the system produced quantitative information on how cisplatin is metabolized and cleared by the liver and kidney, which will make it suitable for more refined predictions of drug absorption, distribution, metabolism, excretion and toxicity.” Herland is now an Associate Professor at KTH Royal Institute of Technology and the Karolinska Institute in Stockholm, Sweden.
Source: Read Full Article